Publications
bioRxiv (2025): A Human Tumor-Immune Organoid Model of Glioblastoma
Shivani Baisiwala, Elisa Fazzari, Matthew X Li, Antoni Martija, Daria J Azizad, Lu Sun, Gilbert Herrera, Trinh Phan, Amber Monteleone, David A Nathanson, Anthony Wang, Won Kim, Richard G Everson, Kunal S Patel, Linda M Liau, Robert M Prins, Aparna Bhaduri
We developed the immune-Human Organoid Tumor Transplantation (iHOTT) model. This is an autologous co-culture platform that integrates patient-derived tumor cells and matched peripheral blood mononuclear cells (PBMCs) within human cortical organoids, enabling the study of the patient-specific immune response to the tumor and tumor-immune interactions. This platform preserves tumor and immune populations, immune signaling, and cell-cell interactions observed in patient tumors. Treatment of iHOTT with pembrolizumab, a checkpoint inhibitor, mirrored cell type shifts and cell interactions observed in patients. TCR sequencing further revealed pembrolizumab-driven expansion of stem-like CD4-T-cell clonotypes exhibiting patient-specific repertoires. These findings establish iHOTT as a physiologically relevant platform for exploring autologous tumor–immune interactions and underscore the critical need for antigen-targeted strategies to enhance immunotherapy in glioblastoma.
bioRxiv (2025): Thalamic NRXN1-Mediated Input to Human Cortical Progenitors Drives Upper Layer Neurogenesis
Claudia V. Nguyen, Antoni Martija, Patricia R. Nano, Jose A. Soto, Daniel C. Jaklic, Jessenya Mil, Rista White, Jacqueline M. Martin, Dakshesh Rana, Daniel H. Geschwind, Aparna Bhaduri
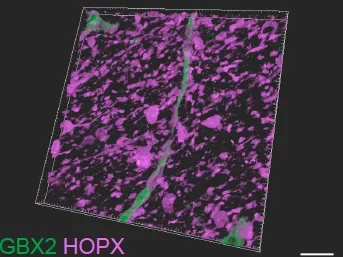
We used the thalamocortical assembloid system to study how thalamic signaling shapes the development and expansion of the developing human cortex. Using single-nuclei RNA-sequencing and cellular imaging, we discover that thalamic signals, during a critical period, promote human cortical upper-layer neurogenesis. In assembloid models and human primary cortex, we find NRXN1 mediates thalamic axon contact with primate-enriched outer radial glia, driving developmental gene expression changes. Genetic perturbation of NRXN1 in thalamic neurons reduces these contacts and attenuates cortical upper-layer neurogenesis. These findings in human developmental models suggest a novel role for thalamic regulation of primate outer radial glia cell fate.
Nature Neuroscience (2025): Integrated analysis of molecular atlases unveils modules driving developmental cell subtype specification in the human cortex
Patricia R Nano, Elisa Fazzari, Daria Azizad, Antoni Martija, Claudia V Nguyen, Sean Wang, Vanna Giang, Ryan L Kan, Juyoun Yoo, Brittney Wick, Maximilian Haeussler, Aparna Bhaduri
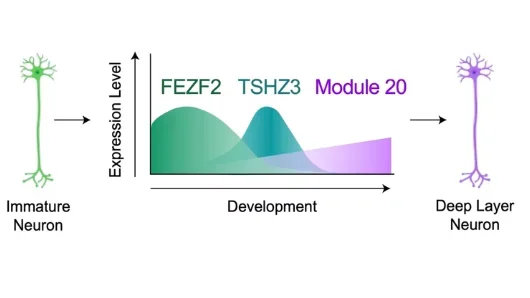
Through parallel meta-analyses of the human cortex in development (seven datasets) and adulthood (16 datasets), we generated over 500 gene co-expression networks that can describe mechanisms of cortical development, centering on peak stages of neurogenesis. These meta-modules show dynamic cell subtype specificities throughout cortical development, with several developmental meta-modules displaying spatiotemporal expression patterns that allude to potential roles in cell fate specification. We validated the expression of these modules in primary human cortical tissues. These include meta-module 20, a module elevated in FEZF2+ deep layer neurons that includes TSHZ3, a transcription factor associated with neurodevelopmental disorders. Human cortical chimeroid experiments validated that both FEZF2 and TSHZ3 are required to drive module 20 activity and deep layer neuron specification but through distinct modalities. These studies demonstrate how meta-atlases can engender further mechanistic analyses of cortical fate specification.
bioRxiv (2025): Metabolic Atlas of Early Human Cortex Identifies Regulators of Cell Fate Transitions
Jessenya Mil, Jose A Soto, Nedas Matulionis, Abigail Krall, Francesca Day, Linsey Stiles, Katrina P Montales, Daria J Azizad, Carlos E Gonzalez, Patricia R Nano, Antoni A Martija, Cesar A Perez-Ramirez, Claudia V Nguyen, Ryan L Kan, Madeline G Andrews, Heather A Christofk, Aparna Bhaduri
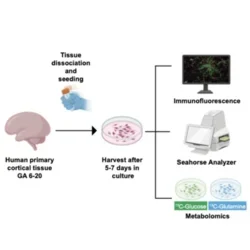
We performed a variety of metabolic assays in primary tissue and stem cell derived cortical organoids and observed dynamic changes in core metabolic functions, including an unexpected increase in glycolysis during late neurogenesis. By depleting glucose levels in cortical organoids, we increased outer radial glia, astrocytes, and inhibitory neurons. We found the pentose phosphate pathway (PPP) was impacted in these experiments and leveraged pharmacological and genetic manipulations to recapitulate these radial glia cell fate changes. These data identify a new role for the PPP in modulating radial glia cell fate specification and generate a resource for future exploration of additional metabolic pathways in human cortical development.
bioRxiv (2024): Glioblastoma Neurovascular Progenitor Orchestrates Tumor Cell Type Diversity
Elisa Fazzari, Daria J Azizad, Kwanha Yu, Weihong Ge, Matthew X Li, Patricia R Nano, Ryan L Kan, Hong A Tum, Christopher Tse, Nicholas A Bayley, Vjola Haka, Dimitri Cadet, Travis Perryman, Jose A Soto, Brittney Wick, David R Raleigh, Elizabeth E Crouch, Kunal S Patel, Linda M Liau, Benjamin Deneen, David A Nathanson, Aparna Bhaduri
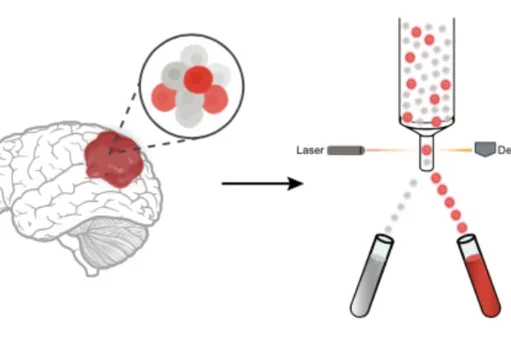
We constructed a gene program-centric meta-atlas of published transcriptomic studies to identify commonalities between diverse GBM tumors and cell types to decipher the mechanisms that drive them. This approach led to the discovery of a tumor-derived stem cell population with mixed vascular and neural stem cell features, termed a neurovascular progenitor (NVP). In-vivo genetic depletion of NVP cells resulted in altered tumor cell composition, fewer cycling cells, and extended survival, underscoring their critical functional role. Clonal analysis of primary patient tumors in a human organoid tumor transplantation system demonstrated that the NVP has dual potency, generating both neuronal and vascular tumor cells. Although NVP cells comprise a small fraction of the tumor, these clonal analyses demonstrated that they strongly contribute to the total number of cycling cells in the tumor and generate a defined subset of the whole tumor. This study represents a paradigm by which cell type-specific interrogation of tumor populations can be used to study functional heterogeneity and therapeutically targetable vulnerabilities of GBM.
bioRxiv (2024): Human-specific paralogs of SRGAP2 induce neotenic features of microglia structural and functional maturation
Carlos Diaz-Salazar, Marine Krzisch, Juyoun Yoo, *Patricia R. Nano*, *Aparna Bhaduri*, Rudolf Jaenisch, Franck Polleux
The authors, using a combination of xenotransplantation of human induced pluripotent stem cell (hiPSC)-derived microglia and mouse genetic models, demonstrate that human-specific SRGAP2B/C paralogs are necessary and sufficient to induce neotenic features of microglia structural and functional maturation in a cell-autonomous manner. Additionally, the induction of SRGAP2-dependent neotenic features of microglia maturation non-cell autonomously impacts synaptic development in cortical pyramidal neurons. The results reveal that, during human brain evolution, human-specific genes SRGAP2B/C coordinated the emergence of neotenic features of synaptic development by acting as genetic modifiers of both neurons and microglia.
Nature News and Views (2024): Chimeric brain organoids capture human genetic diversity
Aparna Bhaduri
Aparna highlights a 2024 publication from Antón-Bolaños et al., describing "Chimeroids": an in-vitro model system that relies on mixing cells from different human donors to create cortical organoids that are genetic chimeras.
bioRxiv (2024): Radiation-Induced Cellular Plasticity: A Strategy for Combatting Glioblastoma
Ling He, *Daria Azizad*, Kruttika Bhat, Angeliki Ioannidis, Carter J. Hoffmann, Evelyn Arambula, *Aparna Bhaduri*, Harley I. Kornblum, Frank Pajonk
The authors used the adenylcyclase activator forskolin to alter the cellular fate of glioma cells in response to radiation. The combined treatment induced the expression of neuronal markers in glioma cells, reduced proliferation and led to a distinct gene expression profile. scRNAseq revealed that the combined treatment forced glioma cells into microglia- and neuron-like phenotypes. In vivo, this treatment led to a loss of glioma stem cells and prolonged median survival in mouse models of glioblastoma. Collectively, the data suggest that revisiting radiation therapy in combination with forskolin could lead to clinical benefit.
bioRxiv (2024): Human Organoid Tumor Transplantation Identifies Functional Glioblastoma - Microenvironmental Communication Mediated by PTPRZ1
Weihong Ge*, Ryan L Kan*, Can Yilgor, Elisa Fazzari, Patricia R Nano, Daria J Azizad, Matthew Li, Joyce Y Ito, Christopher Tse, Hong A Tum, Jessica Scholes, Kunal S Patel, David A Nathanson, Aparna Bhaduri
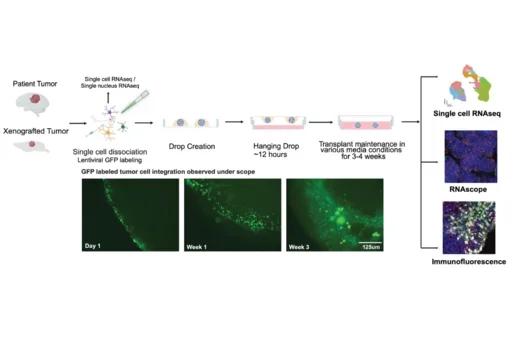
We leveraged our novel human organoid tumor transplantation (HOTT) system to explore how extrinsic cues modulate glioblastoma cell type specification, heterogeneity, and migration. We show that HOTT recapitulates the core features of major patient tumor cell types and key aspects of peritumor cell types, while providing a human microenvironment that uniquely enables perturbations in both the patient tumor and its microenvironment. Our exploration of patient tumor microenvironmental interactions in HOTT highlighted PTPRZ1, a receptor tyrosine phosphatase implicated in tumor migration, as a key player in intercellular communication. We observed that tumor knockdown of PTPRZ1 recapitulated previously described roles in migration and maintaining progenitor identity. Unexpectedly, environmental PTPRZ1 knockdown drove opposite migration and cell fate changes in the tumor, even when the tumor was not manipulated. This previously undiscovered mode of tumor-microenvironmental communication highlights the need to study human glioblastoma in the context of a human microenvironment such as HOTT.
Nature Perspectives (2023): Functional genomics and systems biology in human neuroscience
Genevieve Konopka & Aparna Bhaduri
Aparna and Genevieve outline the progress that has been made in the application of systems-level network analyses to neurogenomics datasets.
Acta Neuropathol Commun (2023): Common molecular features of H3K27M DMGs and PFA ependymomas map to hindbrain developmental pathways
Matthew Pun, Drew Pratt, *Patricia R. Nano*, Piyush K. Joshi, Li Jiang, Bernhard Englinger, Arvind Rao, Marcin Cieslik, Arul M. Chinnaiyan, Kenneth Aldape, Stefan Pfister, Mariella G. Filbin, *Aparna Bhaduri* & Sriram Venneti
The authors identified shared molecular features of H3K27-altered diffuse midline gliomas (DMGs) and group-A posterior fossa ependymomas (PFAs) by comparing genomic, bulk transcriptomic, chromatin-based profiles, and single-cell RNA-sequencing (scRNA-seq) data from the two tumor classes. This approach demonstrated that 1q gain, a key biomarker in PFAs, is prognostic in H3.1K27M, but not H3.3K27M gliomas. Conversely, Activin A Receptor Type 1 (ACVR1), which is associated with mutations in H3.1K27M gliomas, is overexpressed in a subset of PFAs with poor outcomes. Despite diffuse H3K27me3 reduction, previous work shows that both tumors maintain genomic H3K27me3 deposition at select sites. The authors demonstrate heterogeneity in shared patterns of residual H3K27me3 for both tumors that largely segregated with inferred anatomic tumor origins and progenitor populations of tumor cells. In contrast, analysis of genes linked to H3K27 acetylation (H3K27ac)-marked enhancers showed higher expression in astrocytic-like tumor cells. Finally, common H3K27me3-marked genes mapped closely to expression patterns in the human developing hindbrain. Overall, this data demonstrates developmentally relevant molecular similarities between PFAs and H3K27M DMGs and supports the overall hypothesis that deregulated mechanisms of hindbrain development are central to the biology of both tumors.
Dev Neurobio (2022): Evaluation of advances in cortical development using model systems
Patricia R. Nano, Aparna Bhaduri
The Bhaduri Lab reviews key stages of cortical development and highlights known or possible differences between model systems and the developing human brain. By identifying the developmental trajectories that may facilitate uniquely human traits, we highlight open questions in need of approaches to examine these processes in a human context and reveal translatable insights into human developmental disorders.
Frontiers (2021): Cortical Cartography: Mapping Arealization Using Single-Cell Omics Technology
Patricia R. Nano, Claudia V. Nguyen, Jessenya Mil, Aparna Bhaduri
The Bhaduri Lab reviews the single-omics atlases that have shaped our current understanding of cortical areas, and their potential to fuel a new era of multi-omic single-cell endeavors to interrogate both the developing and adult human cortex.
Nature (2021): An atlas of cortical arealization identifies dynamic molecular signatures
Aparna Bhaduri, Carmen Sandoval-Espinosa, Marcos Otero-Garcia, Irene Oh, Raymund Yin, Ugomma C. Eze, Tomasz J. Nowakowski & Arnold R. Kriegstein
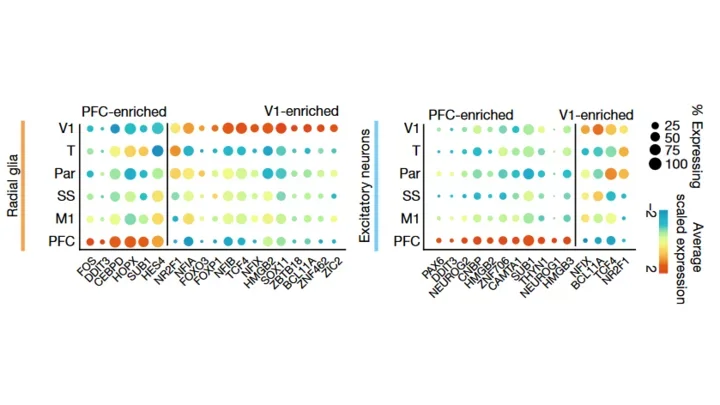
We used single-cell RNA sequencing to profile ten major brain structures and six neocortical areas during peak neurogenesis and early gliogenesis. Within the neocortex, we find that early in the second trimester, a large number of genes are differentially expressed across distinct cortical areas in all cell types, including radial glia, the neural progenitors of the cortex. However, the abundance of areal transcriptomic signatures increases as radial glia differentiate into intermediate progenitor cells and ultimately give rise to excitatory neurons. Using an automated, multiplexed single-molecule fluorescent in situ hybridization approach, we find that laminar gene-expression patterns are highly dynamic across cortical regions. Together, our data suggest that early cortical areal patterning is defined by strong, mutually exclusive frontal and occipital gene-expression signatures, with resulting gradients giving rise to the specification of areas between these two poles throughout successive developmental timepoints.
Nature Neuroscience (2021): Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia
Ugomma C. Eze*, Aparna Bhaduri*, Maximilian Haeussler, Tomasz J. Nowakowski and Arnold R. Kriegstein
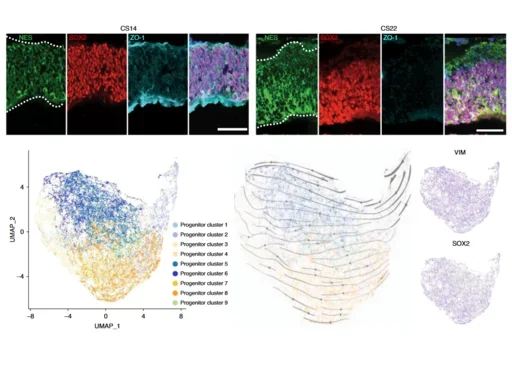
We performed single-cell RNA-sequencing across regions of the developing human brain, including the telencephalon, diencephalon, midbrain, hindbrain and cerebellum. We identified nine progenitor populations physically proximal to the telencephalon, suggesting more heterogeneity than previously described, including a highly prevalent mesenchymal-like population that disappears once neurogenesis begins. Comparison of human and mouse progenitor populations at corresponding stages identifies two progenitor clusters that are enriched in the early stages of human cortical development. We also find that organoid systems display low fidelity to neuroepithelial and early radial glia cell types, but improve as neurogenesis progresses. Overall, we provide a comprehensive molecular and spatial atlas of early stages of human brain and cortical development.
Cell (2020): Origins and Proliferative States of Human Oligodendrocyte Precursor Cells
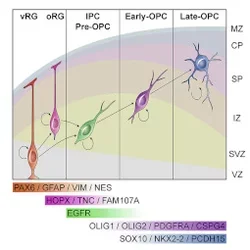
Huang H*, Bhaduri A*, Velmeshev D, Wang S, Wang L, Rottkamp CA, Alvarez-Buylla A, Rowitch DH, Kriegstein AR.
Using single-cell RNA sequencing of immunopanned oligodendrocyte precursor cells (OPC), we identified a novel pre-OPC population that is generated from outer radial glia cells and is marked by EGFR. We show that this population requires EGFR mediated signaling to produce oligodendrocytes. Because this pre-OPC cell type comes from outer radial glia cells which are enriched in humans, these cells may play a role in cortical expansion.
Nature (2020): Cell stress in cortical organoids impairs molecular subtype specification
Bhaduri A*, Andrews MG*, Jung D, Shin D, Allen D, Schmunk G, Pollen AA, Nowakowski TJ, Kriegstein AR.
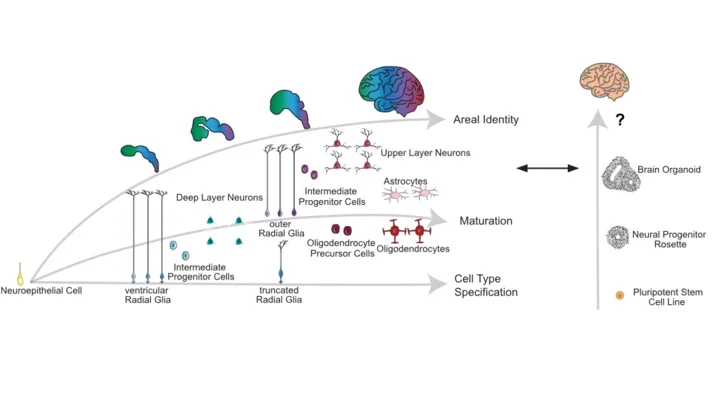
We compared cell types in cortical organoid systems and the primary developing human brain using single-cell RNA sequencing. We identify that major cell types are preserved across both systems, but that cortical organoids suffer from impaired cell subtype specification, incongruent maturation, and areal heterogeneity. We observe that organoids have higher levels of metabolic stress and that activation of this stress in primary cells in culture also impairs subtype specification, while transplantation of organoids into the mouse brain alleviates stress and improves subtypes. These data suggest that organoid cultures can be improved to be more physiologically relevant, but also demonstrates that the limitations of cell type specification can be resolved.
Cell Stem Cell (2020): Outer Radial Glia-like Cancer Stem Cells Contribute to Heterogeneity of Glioblastoma
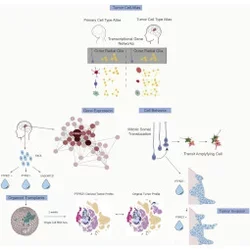
Bhaduri A*, Di Lullo E*, Jung D, Müller S, Crouch EE, Sandoval Espinosa C, Ozawa T, Alvarado B, Spatazza J, Cadwell CR, Wilkins G, Velmeshev D, Liu SJ, Malatesta M, Andrews MG, Mostajo-Radji MA, Huang EJ, Lim DA, Nowakowski TJ, Diaz A, Raleigh DR, Kriegstein AR.
To understand the developmental origins of glioblastoma, we collected single-cell RNA sequencing from 11 primary tumors and compared it to our developmental datasets. We identified the reactivation of the outer radial glia cell, and observed it was undergoing its classic "jump and divide" mitotic somal translocation behavior that may be mediated tumor invasion via PTPRZ1. Using a novel tumor transplantation assay into cortical organoids, we saw these cells were capable of giving rise to neuronal and glial tumor populations.
Cell (2019): Establishing Cerebral Organoids as Models of Human-Specific Evolution
Pollen AA*, Bhaduri A*, Andrews MG, Nowakowski TJ, Meyerson O, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty M, Fiddes I, Kronenberg Z, Shuga J, Leyrat A, West J, Bershteyn M, Lowe C, Pavlovic B, Salama SR, Haussler D, Eichler EE, Kriegstein AR.
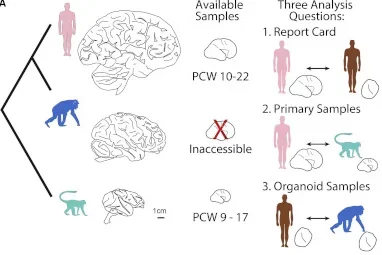
The human cortex is substantially expanded compared to our closest relatives, and these differences emerge at birth. Using primary human and macaque developing cortex, as well as cortical organoids from human and chimpanzee cells, we probed these differences using single-cell RNA sequencing. We identified major cell populations conserved across all systems, and highlight several hundred genes that are differentially expressed between human and non-human primates. These genes highlighted mTOR signaling to be up-regulated in the human cortex, and we find this activation is specific to the outer radial glia cells, suggesting cell type specific pathway activation may contribute to cortical expansion.
Science (2017): Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex
Nowakowski TJ*, Bhaduri A*, Pollen AA*, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, Ounadjela JR, Shuga J, Wang X, Lim DA, West JA, Leyrat AA, Kent WJ, Kriegstein AR.
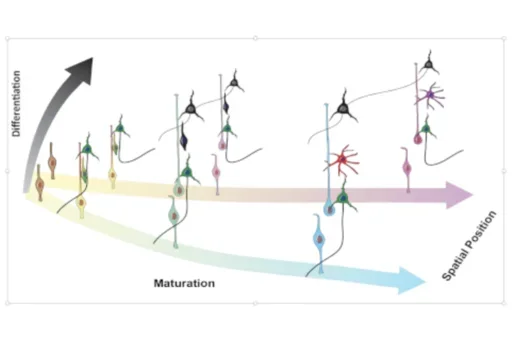
The human cortex is substantially expanded compared to our closest relatives, and these differences emerge at birth. Using primary human and macaque developing cortex, as well as cortical organoids from human and chimpanzee cells, we probed these differences using single-cell RNA sequencing. We identified major cell populations conserved across all systems, and highlight several hundred genes that are differentially expressed between human and non-human primates. These genes highlighted mTOR signaling to be up-regulated in the human cortex, and we find this activation is specific to the outer radial glia cells, suggesting cell type specific pathway activation may contribute to cortical expansion.